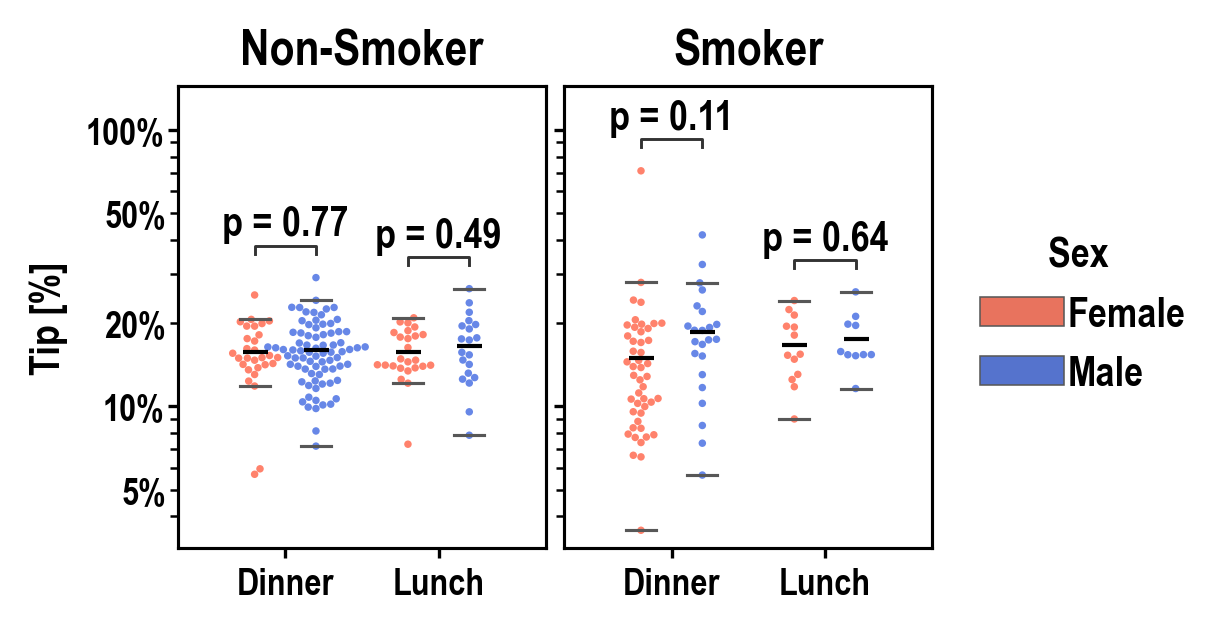
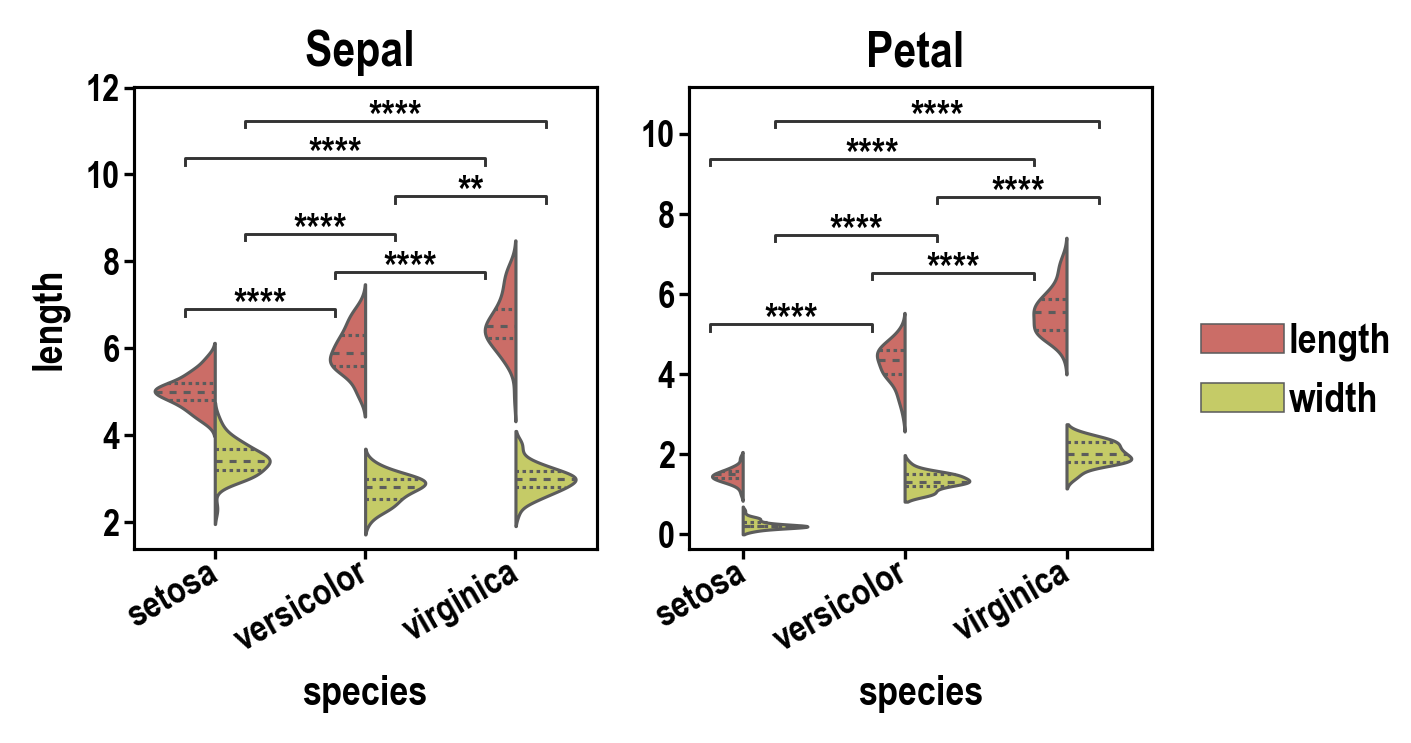
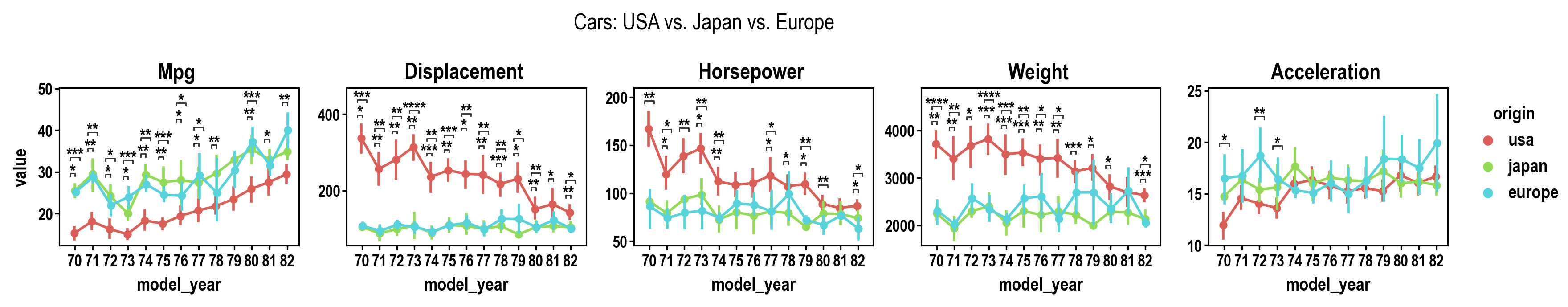
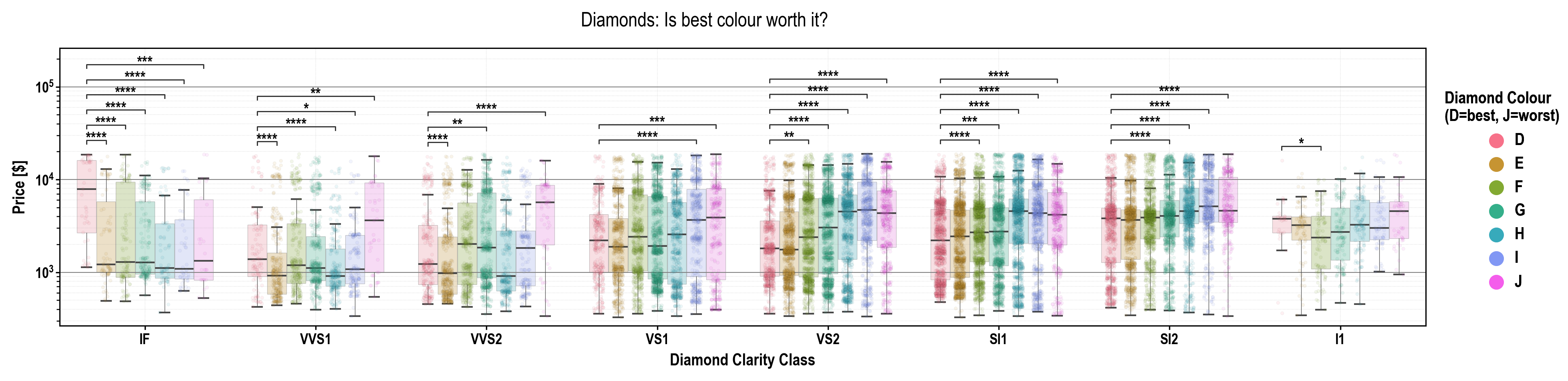
pip install plotasticpip install git+https://github.com/markur4/plotastic.git- Python >= 3.11 (not tested with earlier versions)
- pandas == 1.5.3 (pingouin needs this)
- seaborn <= 0.12.2 (later versions reworked hue)
🤔 Summary
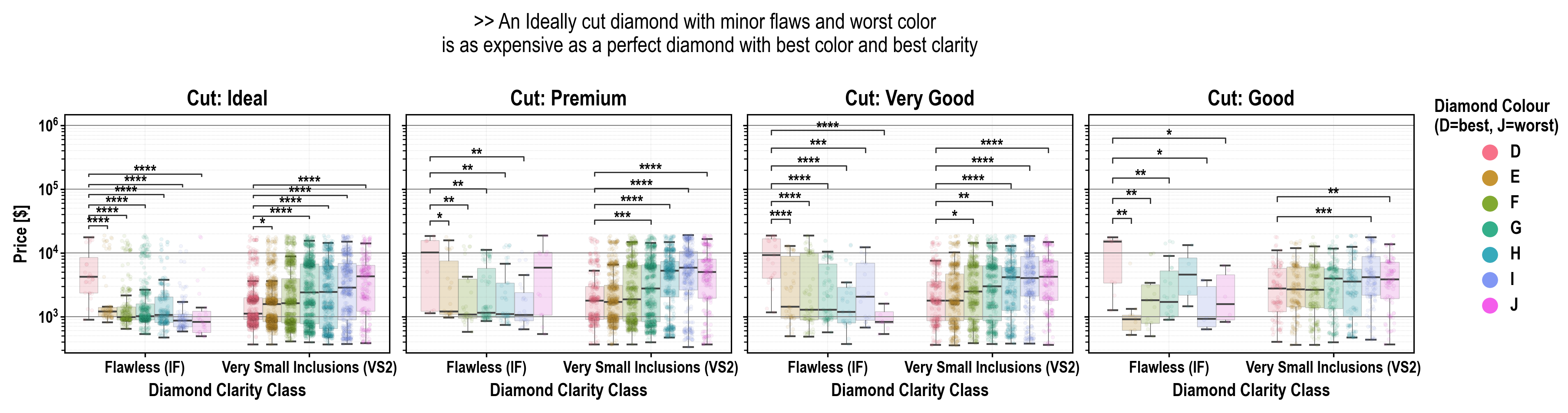
plotasticaddresses the challenges of transitioning from exploratory data analysis to hypothesis testing in Python's data science ecosystem. Bridging the gap betweenseabornandpingouin, this library offers a unified environment for plotting and statistical analysis. It simplifies the workflow with a user-friendly syntax and seamless integration with familiarseabornparameters (y, x, hue, row, col). Inspired byseaborn's consistency,plotasticutilizes aDataAnalysisobject to intelligently pass parameters topingouinstatistical functions. The library systematically groups the data according to the needs of statistical tests and plots, conducts visualisation, analyses and supports extensive customization options. In essence,plotasticestablishes a protocol for configuring statical analyses through plotting parameters. This approach streamlines the process, translatingseabornparameters into statistical terms, providing researchers and data scientists with a cohesive and user-friendly solution in python.!
- 🧮 Import & Prepare your pandas DataFrame
- We require a long-format pandas dataframe with categorical columns
- If it works with seaborn, it works with plotastic!
- 🔀 Make a DataAnalysis Object
DataAnalysis(DataFrame, dims={x, y, hue, row, col})- Check for empty data groups, differing samplesizes, NaN-count, etc. automatically
- ✅ Explore Data
- Check Data integrity, unequal samplesizes, empty groups, etc.
- Quick preliminary plotting with e.g.
DataAnalysis.catplot()- 🔨 Adapt Data
- Categorize multiple columns at once
- Transform dependent variable
- Each step warns you, if you introduced NaNs without knowledge!
- etc.
- ✨ Perform Statistical Tests ✨
- Check Normality, Homoscedasticity, Sphericity
- Perform Omnibus tests (ANOVA, RMANOVA, Kruskal-Wallis, Friedman)
- Perform PostHoc tests (Tukey, Dunn, Wilcoxon, etc.) based on
pg.pairwise_tests()- 📊 Plot figure
- Use pre-defined and optimized multi-layered plots with one line (e.g. strip over box)!
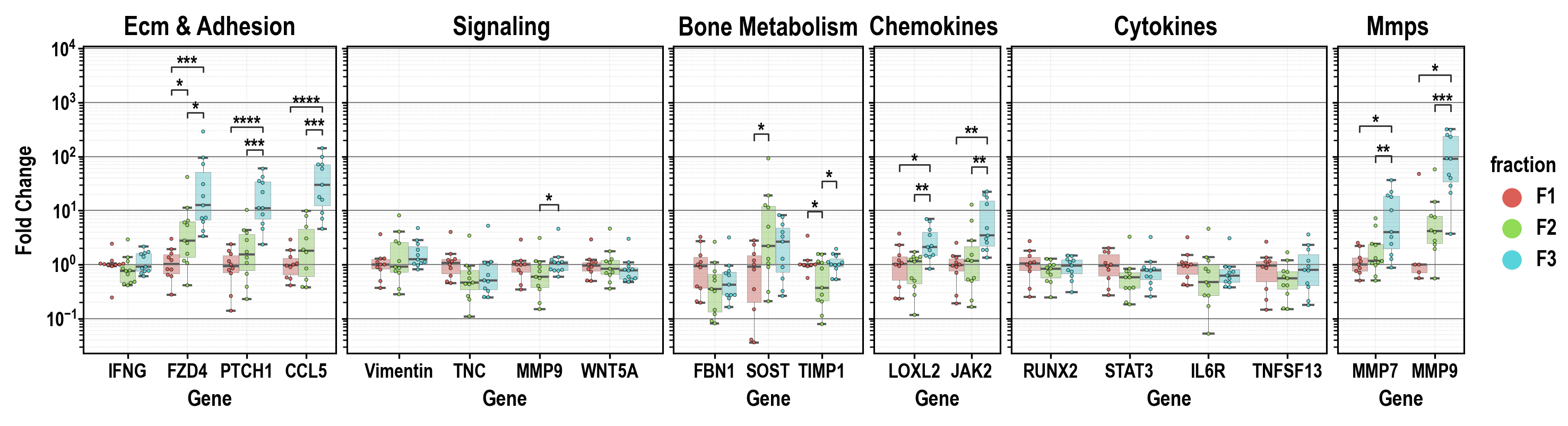
- Annotate statistical results (p-values as *, **, ***, etc.) with full control over which data to include or exclude!
- 💿 Save all results at once!
- One DataAnalysis object holds:
- One DataFrame in
self.data- One Figure in
self.fig,self.axes- Multiple statistical results:
self.results- Use
DataAnalysis.save_statistics()to save all results to different sheets collected in one .xlsx filesheet per test
📊 Translating Plots into Statistics!
- Categorical data is separable into
seaborn's categorization parameters: x, y, hue, row, col. We call those "dimensions".- These dimensions are assigned to statistical terms:
- y is the dependent variable (DV)
- x and hue are independent variables (IV) and are treated as within/between factors (categorical variables)
- row and col are grouping variables (categorical variables)
- A subject may be specified for within/paired study designs (categorical variable)
- For each level of row or col (or for each combination of row- and col levels), statistical tests will be performed with regards to the two-factors x and hue
- Imagine this example data:
- Each day you measure the tip of a group of people.
- For each tip, you note down the day, gender, age-group and whether they smoke or not.
- Hence, this data has 4 categorical dimensions, each with 2 or more levels:
- day: 4 levels (monday, tuesday, wednesday, Thursday)
- gender: 2 levels (male, female)
- smoker: 2 levels (yes, no)
- age-group: 2 levels (young, old)
- Each category is assigned to a place of a plot, and when calling statistical tests, we assign them to statistical terms (in comments):
- We perform statistical testing groupwise:
- For each level-combinations of smoker and age-group, a two-way ANOVA will be performed (with day and gender as between factors for each datagroup):
- 1st ANOVA assesses datapoints where smoker=yes AND age-group=young
- 2nd ANOVA assesses datapoints where smoker=yes AND age-group=old
- 3rd ANOVA assesses datapoints where smoker=no AND age-group=young
- 4th ANOVA assesses datapoints where smoker=no AND age-group=old
- Three-way ANOVAs are not possible (yet), since that would require setting e.g. col as the third factor, or implementing another dimension (e.g. hue2).
❗️ Disclaimer about Statistics
- ... Intuitive Biostatistics - Fourth Edition (2017); Harvey Motulsky
- ... Introduction to Statistical Learning with applications in Python - First Edition (2023); Gareth James, Daniela Witten, Trevor Hastie, Robert Tibshirani, Jonathan Taylor
- ... talking to other scientists struggling with statistics
- ... gaining some practical experience when learning statistics
- ... quickly gain statistical implications about your data without switching to another software
- ... making first steps towards a full statistical analysis
- ... plotting publication grade figures (check statistics results with other software)
- ... publication grade statistical analysis IF you really know what you're doing OR you have back-checked your results by a professional statistician
- ... quickly test data transformations (log)
- ... replace a professional statistician
- ... teach you statistics, you need some basic knowledge (but is awesome for practicing!)
- ... test for multicolinearity (Absence of multicolinearity is required by ANOVA!)
- ... perform stringent correction for multiple testing (e.g. bonferoni), as statistical tests are applied to sub-facets of the whole dataframe for each axes, which depends on the definition of x, hue, col, etc. Hence, corrected p-values might over-estimate the significance of your results.
- Expect Errors: Don't trust automated systems like this one!
- Document your work in ridiculous detail:
- Include the applied tests, the number of technical replicates and the number of biological/independent in each figure legend
- State explicitly what each datapoint represents:
- 1 datapoint = 1 Technical replicate?
- 1 datapoint = The mean of all technical replicate per independent replicate/subject?
- State explicitly what the error-bars mean: Standard deviation? Confidence interval?
- (Don't mix technical with biological/independent variance)
- Report if/how you removed outliers
- Report if you did or did not apply correction methods (multiple comparisons, Greenhouse Geyser, etc.) and what your rationale is (exploratory vs. confirmatory study? Validation through other methods to reduce Type I error?)
- Check results with professionnals:
- "Here is my data, here is my question, here is my analysis, here is my interpretation. What do you think?"
✅ Feature List
- ✅: Complete and tested
- 👍: Complete
- 📆: Planned or unfinished (no date)
- 🤷: Maybe..? (Rather not...)
- 🚫: Not planned, don't want
- 😣: Help Please..?
Plotting
- 👍 Make and Edit Plots: Implemented ✅
- All (non-facetgrid) seaborn plots should work, not tested
- 📆 QQ-Plot
- 📆 Kaplan-Meyer-Plot
- 🤷 Interactive Plots (where you click stuff and adjust scale etc.)
- That's gonna be alot of work!
- 🚫 Support for
seaborn.FacetGrid
- Why not? -
plotasticuses matplotlib figures and fills its axes with seaborn plot functions. In my opinion, that's the best solution that offers the best adaptibility of every plot detail while bieng easy to maintain- 🚫 Support for
seaborn.objects(same as Facetgrid)
- Why not? - I don't see the need to refactor the code
- 😣 NEED HELP WITH: The hidden state of
matplotlibfigures/plots/stuff that gets drawn:
- I want to save the figure in
DataAnalysis.figattribute. As simple as that sounds, matplotlib does weird stuff, not applying changes after editing the plot.- It'd be cool if I could control the changes to a DataAnalysis object better (e.g. using
inplace=Truelike withpd.DataFrames). But I never figured out how to control matplotlib figure generation, even with re-drawing the figure with canvas. It's a mess and I wasted so much time already.Multi-Layered Plotting
- ✅ Box-plot + swarm
- 👍 Box-plot + strip
- 📆 Violin + swarm/strip
Statistics
- Assumption testing
- ✅ Normality (e.g. Shapiro-Wilk)
- ✅ Homoscedasticity (e.g. Levene)
- ✅ Sphericity (e.g. Mauchly)
- Omnibus tests
- ✅ ANOVA, RMANOVA, Kruskal-Wallis, Friedman
- 📆 Mixed ANOVA
- 📆 Annotate Results into Plot
- PostHoc
- ✅
pg.pairwise_tests()
- Works with all primary options. That includes all parametric, non-parametric, paired, unpaired, etc. tests (t-test, paired t-test, MWU, Wilcoxon, etc.)
- ✅ Annotate Stars into plots (*, **, etc.)
- Specific pairs can be included/excluded from annotation
- 📆 Make correction for multiple testing go over complete DataFrame and not Facet-wise:
- Bivariate
- 📆 Find and Implement system to switch between numerical and categorical x-axis
- Function to convert numerical data into categorical data by binning?
- 📆 Pearson, Spearman, Kendall
Analysis Pipelines
Idea: Put all those statistical tests into one line. I might work on this only after everything's implemented and working confidently and well!
- 🤷
between_samples(parametric=True): ANOVA + Tukey (if Normality & Homoscedasticity are given)- 🤷
between_samples(parametric=False): Kruskal-Wallis + Dunn- 🤷
within_samples(parametric=True): RM-ANOVA + multiple paired t-tests (if Normality & Sphericity are given)- 🤷
within_samples(parametric=False): Friedman + multiple Wilcoxon
🌳 Class Diagram
- 🛑 Not everything shown here is implemented and not everything that's implemented is shown here!
- 🐁 Open the raw .svg in your browser and click on a class to see its source code!

- Example Gallery
- Data
- Plotting
import matplotlib.pyplot as plt
import plotastic as plst
# Import Example Data (Long-Format)
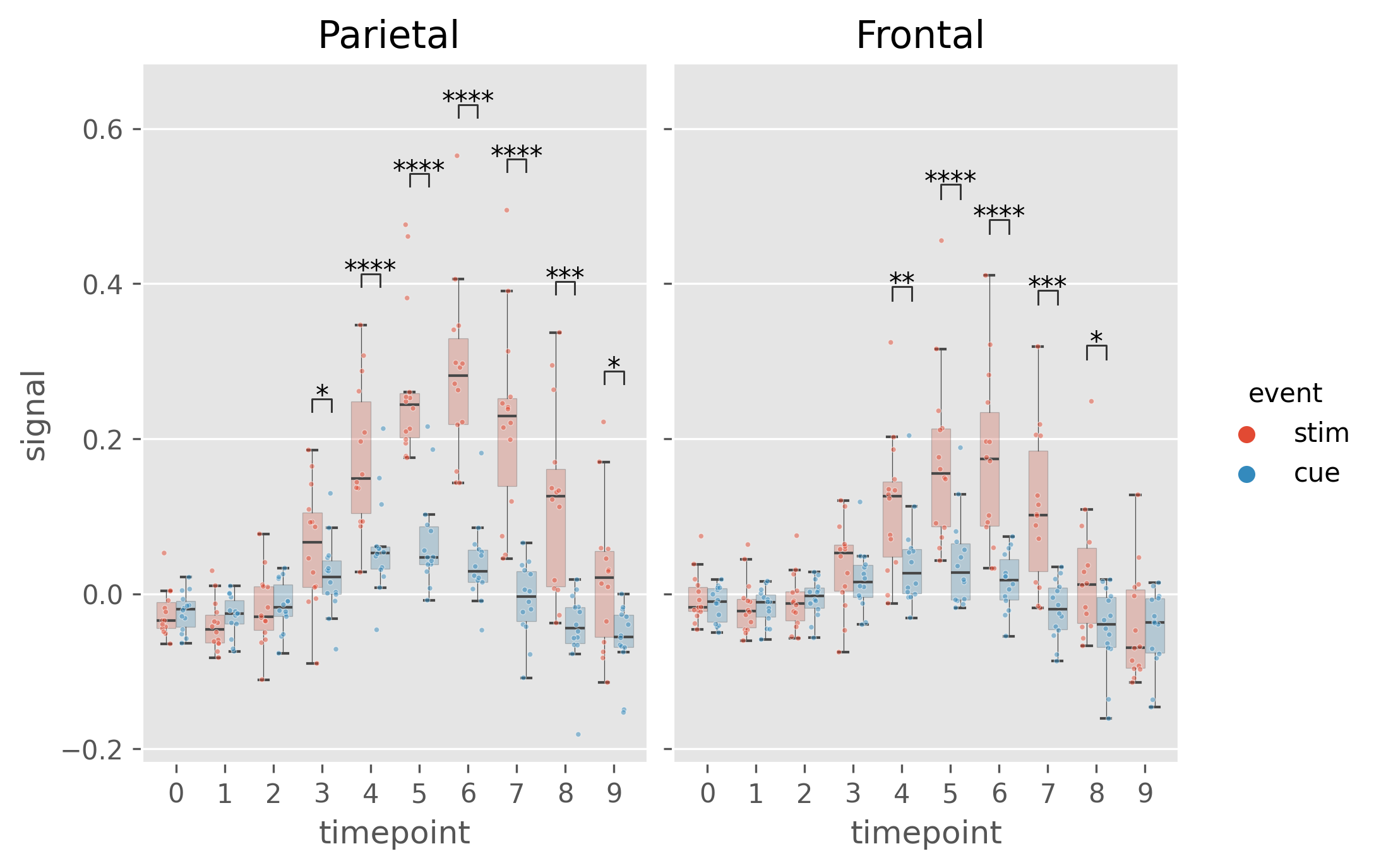
DF, _dims = plst.load_dataset("fmri", verbose = False)
DF.head()| subject | timepoint | event | region | signal | |
|---|---|---|---|---|---|
| 0 | s7 | 9 | stim | parietal | 0.059 |
| 1 | s8 | 9 | stim | parietal | 0.17 |
| 2 | s0 | 0 | stim | frontal | -0.021 |
| 3 | s1 | 0 | stim | parietal | -0.064 |
| 4 | s13 | 9 | stim | parietal | 0.013 |
dims = dict(
y = "signal", # y-axis, dependent variable
x = "timepoint", # x-axis, independent variable & within-subject factor
hue = "event", # color, grouping variable & within-subject factor
col = "region" # axes, grouping variable
)DA = plst.DataAnalysis(
data=DF, # Dataframe, long format
dims=dims, # Dictionary with y, x, hue, col, row
subject="subject", # Datapoints are paired by subject (optional)
verbose=False, # Print out info about the Data (optional)
)No arguments need to be passed, although **kwargs, are passed
to respective pingouin functions.
DA.check_normality() # Normal Distribution?
DA.check_sphericity() # Sphericity?
DA.omnibus_rm_anova() # Repeated Measures ANOVA
DA.test_pairwise() # Post-hoc testsOutput is one excel file containing results of all performed tests (normality, anova, t-tests, etc.) in different sheets
DA.save_statistics("example.xlsx")(DA
.plot_box_strip() # Pre-built plotting function initializes plot
.annotate_pairwise( # Annotate results from DA.test_pairwise()
include="__HUE" # Use only significant pairs across each hue
)
)
# Saving the plot like matplotlib!
plt.savefig("example.png", dpi=200, bbox_inches="tight")(click to unfold)
- Download/Clone repository
- Install development tools
pip install .[dev]- Run tests
- Run
pytest ./tests- To include a coverage report run
pytest ./tests -cov--cov-report=htmland open./htmlcov/index.htmlwith your browser.
(click to unfold)
When interacting with the community, you must adhere to the Code of Conduct
I am grateful for pull requests!
- Make sure to understand the code (e.g. see Class diagram in this Readme)
- Run tests before submitting a pull request
If you need help, please open an issue on this repository.
- Please provide a minimal example to reproduce the problem.
If you need help, please open an issue on this repository.
(click to unfold)
Kuric et al., (2024). plotastic: Bridging Plotting and Statistics in Python. Journal of Open Source Software, 9(95), 6304, https://doi.org/10.21105/joss.06304
Vallat, R. (2018). Pingouin: statistics in Python. Journal of Open Source Software, 3(31), 1026, https://doi.org/10.21105/joss.01026
Waskom, M. L., (2021). seaborn: statistical data visualization. Journal of Open Source Software, 6(60), 3021, https://doi.org/10.21105/joss.03021.
@article{Kuric2024, doi = {10.21105/joss.06304}, url = {https://doi.org/10.21105/joss.06304}, year = {2024}, publisher = {The Open Journal}, volume = {9}, number = {95}, pages = {6304}, author = {Martin Kuric and Regina Ebert}, title = {plotastic: Bridging Plotting and Statistics in Python}, journal = {Journal of Open Source Software} } @article{Waskom2021, doi = {10.21105/joss.03021}, url = {https://doi.org/10.21105/joss.03021}, year = {2021}, publisher = {The Open Journal}, volume = {6}, number = {60}, pages = {3021}, author = {Michael L. Waskom}, title = {seaborn: statistical data visualization}, journal = {Journal of Open Source Software} } @article{Vallat2018, title = "Pingouin: statistics in Python", author = "Vallat, Raphael", journal = "The Journal of Open Source Software", volume = 3, number = 31, pages = "1026", month = nov, year = 2018 }