Replies: 3 comments 19 replies
-
|
There's been some work done during the ICESat-2 Hackweek 2023 to benchmark reading ICESat-2 ATL03 data from an s3 bucket using different libraries and file formats. See:
Preliminary results from ICESAT-2HackWeek/h5cloud@1f34411: Observations:
Caveats: Note that the above ATL03 dataset is a non-gridded point-cloud-like dataset, and may not apply to other HDF5 data structures, so be careful when generalizing this to all HDF5 data files. Next stepsSome of these are outlined by @abarciauskas-bgse at ICESAT-2HackWeek/h5cloud#18:
|
Beta Was this translation helpful? Give feedback.
-
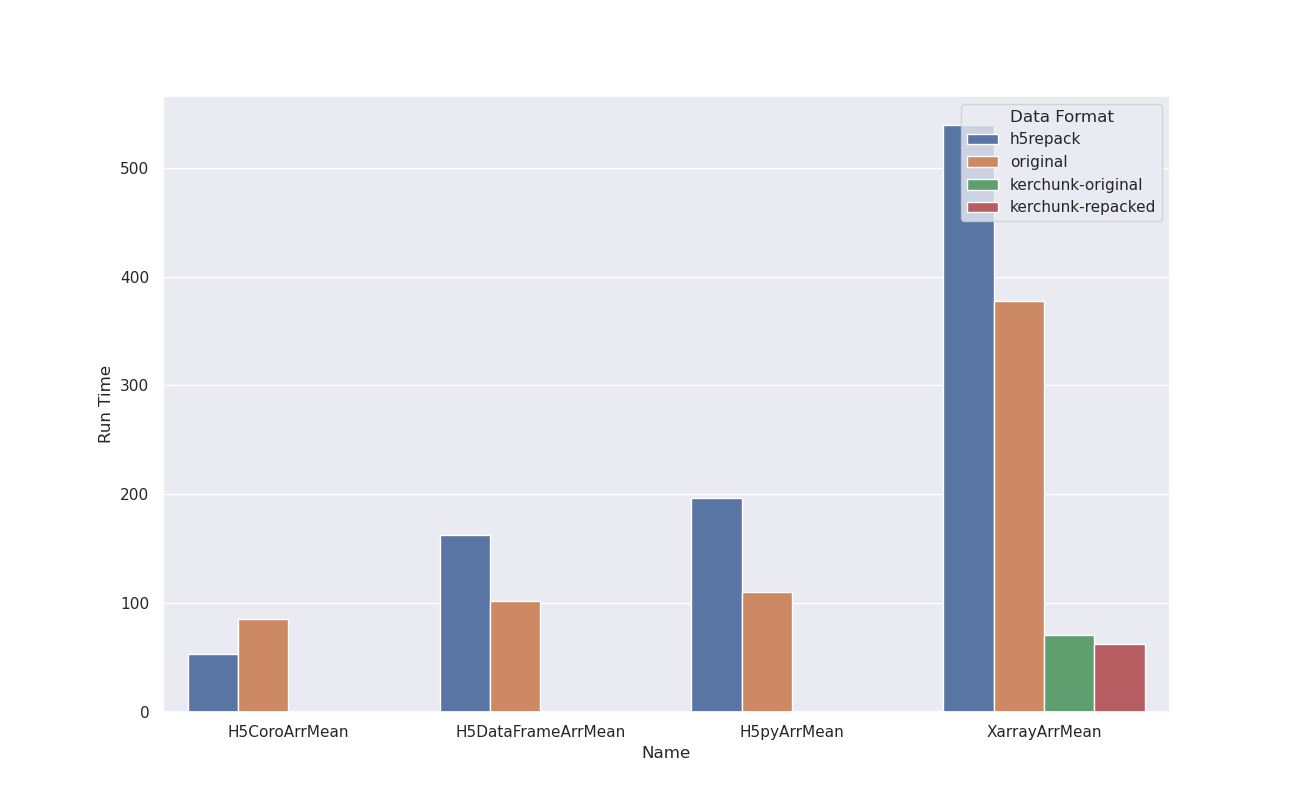
Cloud optimized HDF5 works!Well, seems like cloud-optimized HDF5 is indeed possible and the mixed numbers we initially got were the result of not understanding the default IO and caching behavior of There are some caveats, Exhibit A)Using h5py to calculate a mean (~47000000 data points) in a 7GB file in-region (us-west-2) Observations
Exhibit B)Doing the same operation using Xarray, in-region access on the same 7GB file Observations
Now let's see what happens when we try to do the same operation over 2 files but this time out of region... Exhibit C)Out of region access Observations
We are working on a set of recommendations specifically for ICESat-2 data and we'll share more findings once we have the document ready. |
Beta Was this translation helpful? Give feedback.
-
|
Hi, Great results! Have you attempted to use xarray and dask with the processes scheduler? I'm experimenting with this, but for some unknown reason, the volume of data (Total Req) is huge, leading to longer processing times. For instance: import xarray
import dask
import fsspec
dask.config.set(scheduler="processes")
URL = "..."
fs = fsspec.open(URL, cache_type="blockcache", block_size=8*1024*1024)
ds = xarray.open_dataset(
fs.open(),
engine="h5netcdf",
driver_kwds={
"page_buf_size": 32*1024*1024,
"rdcc_nbytes": 8*1024*1024
}).chunk({...}) # align with file chunking
ds["variable"].mean().compute(num_workers=8) |
Beta Was this translation helpful? Give feedback.



-
Based on the latest discussions on Openscapes about HDF in the cloud, I think there should be a more in-depth study of the state of things, in a way a "follow up" to this post from Matt Rocklin https://matthewrocklin.com/blog/work/2018/02/06/hdf-in-the-cloud
earthaccessusesfsspecfor IO but the HDF libraries usually used to read these files(h5py,pyhdf) were not designed for concurrent access via python file-like objects and IO latency for data in the cloud is really problematic. Here is a list of some of the access patterns used for HDF data in remote storage systems:earthaccessuses, it works but is really slow(not necessarily fsspec's fault), see: h5py sequential reads in the cloud it may also run into deadlock issues when multiple files are accessed.no stand-alone client librarynew development!!, H5Coro has a stand alone client and could be a great drop-in replacement for H5Py on certain casesIdeally we could come up with a better understanding of these access patterns: their capabilities, bottlenecks and potential workarounds.
Beta Was this translation helpful? Give feedback.
All reactions