Tired of generating primers by guessing and trying to predict which nucleotide sequence fits the requirements for PCR/qPCR? This was extremely time-consuming because I had to do this for every gene I wanted to investigate.
To make my PhD research easier, I created a Python program that will tell you whether your primer sequence meets the criteria for efficiency.
You will need to download this folder, and in command line, route to this folder using cd command and run this script.
git clone https://github.com/ying-li-python/DNA-analyze.git
cd DNA-analyze
python main.py
The script will run and will ask you to paste a DNA sequence.
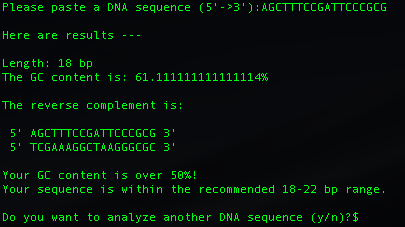
Please paste a DNA sequence (5'->3'):
After printing the results, script will automatically asks the user if you would like to analyze another DNA sequence.
If your DNA sequence contains a non-DNA letter, an error will pop-up and the script will not run.
This is not a valid DNA sequence! Please try again.
We would like the script to show:
- %GC content
- Sequence length
- Reverse complemntary sequence (5' -> 3')
For this script, we created nested for-loops and while loops, if else statements, .replace() and len() function.
- Ying Li
- Python for Biologists by Dr. Martin Jones